Fellow @ JGI
Current Work (2019-2020)
Climate change is an alarming and immediate concern and it is essential to examine myriad facets contributing to the emission of greenhouse gases. Data are needed as input for predictive models to determine outcomes of global change. There is well characterized dynamic carbon flux at interfaces between aquatic and terrestrial systems (McClain et al., 2003, Battin et al., 2009). Engineered transitional systems, such as aqueducts and networks of irrigation canals, have yet to be investigated for carbon flux. There is evidence that some of California’s San Joaquin Delta wetland restorations are contributing methane to the atmosphere (Tringe et al., in progress). The San Joaquin Delta wetlands provide input water for several watersheds and much of this water is pumped into the Central Valley for agriculture. Human-made waterway sediments likely contain microorganismal communities that impact carbon gas flux; mapping the biogeographical distribution of organisms and defining their functional potential across the water distribution system will provide input data for predictive models of landscape features not currently reflected in climate models.
The Delta-Mendota aqueduct is 117 miles long and spans five counties in California (USGS). There are 723 miles of canals within the 154,000 total acres of the Merced Irrigation District Watershed (CA Water Board, 2013). These terrestrial aquatic interfaces cover large regions and are not currently being routinely measured for carbon flux and the microbial communities of these waterways and their potential contribution of greenhouse gases to the atmosphere has yet to be investigated. These engineered interfaces may act as “hotspots” with impacts disproportionate to their geographic area and there is evidence that this is the case in other systems (Wilkins, et al., 2017). Elucidation of the community structure of these large scale engineered terrestrial aquatic habitats may determine if these interfaces require monitoring. this research begins the important work of determining the potential impacts of the microbial community activity on biogeochemical processes occurring in these engineered waterways.
PhD thesis
“Biopesticides” are classified non-pathogenic and classified as Generally Regarded As Safe (GRAS) by the USDA. Considered “green,” “organic,” and safe for consumption, microbial products offer welcome alternatives to pesticides known to damage human health and the environment. In California, close to 8 million pounds of these products are distributed annually. Agricultural practices have been implicated in the spread and generation of antibiotic resistant bacteria. There is an urgent need to understand the role bacterial products play in the spread of antibiotic resistance and novel pathogenicity generation. Addition of biopesticides containing clinically relevant antibiotic resistance genes (ARG) may be creating increased opportunities for exchange. I complied a database of available biopesticide genomes used in CA industrial agriculture and sequenced commercial B. thuringiensis(Bt). Sequences were queried against CARD, ResFinder, and a curated plasmid database. My analysis shows that all of the analyzed biopesticide strains with ARG contain at least one antibiotic drug class and 60% contained four or more drug classes. The main resistance mechanism for both mined and sequenced Bt genomes was antibiotic inactivation.
There was overlap in the resistome species composition from the databases, yet the plasmid database reveals a wider picture of the resistome by including opportunistic pathogens. Bt strains, mined and sequenced, contained diverse ARGs and numerous plasmids of primary pathogens. These results suggest inclusion of plasmids with individual ARG analysis provides a more robust, and likely necessary, view of resistomes. This work indicates that currently used biopesticides contain clinically relevant antibiotic resistance genes, bear resistance to multiple drug classes, and are potential vectors of unintended transmission of antibiotic resistance via plasmid exchange as they are introduced to the environment in large quantities. I also looked at the location of ARG using Hi-C, viral content, and vector behavior using co-culture experiments with known antibiotic resistant pathogenic strains.
URM Ugrad Research
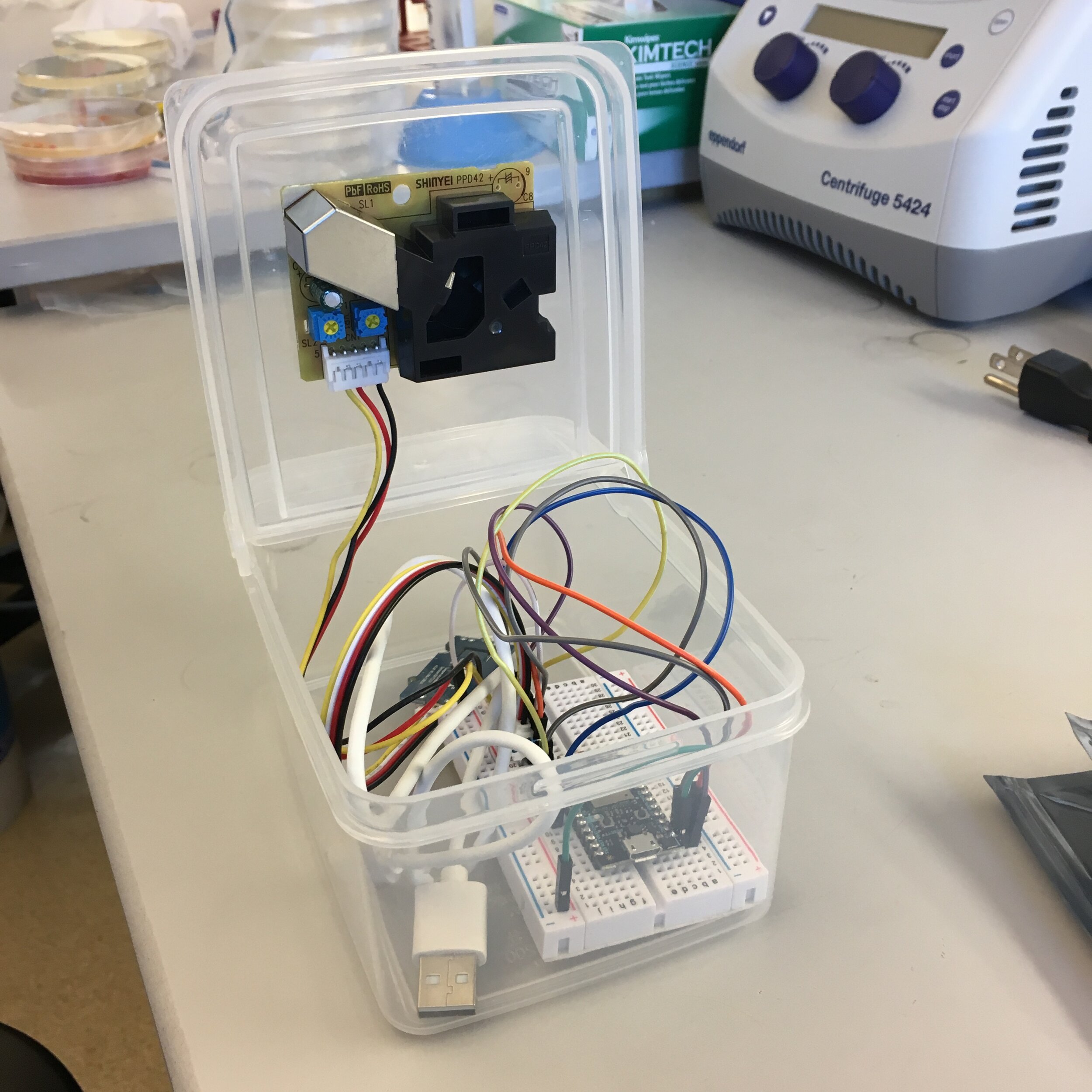
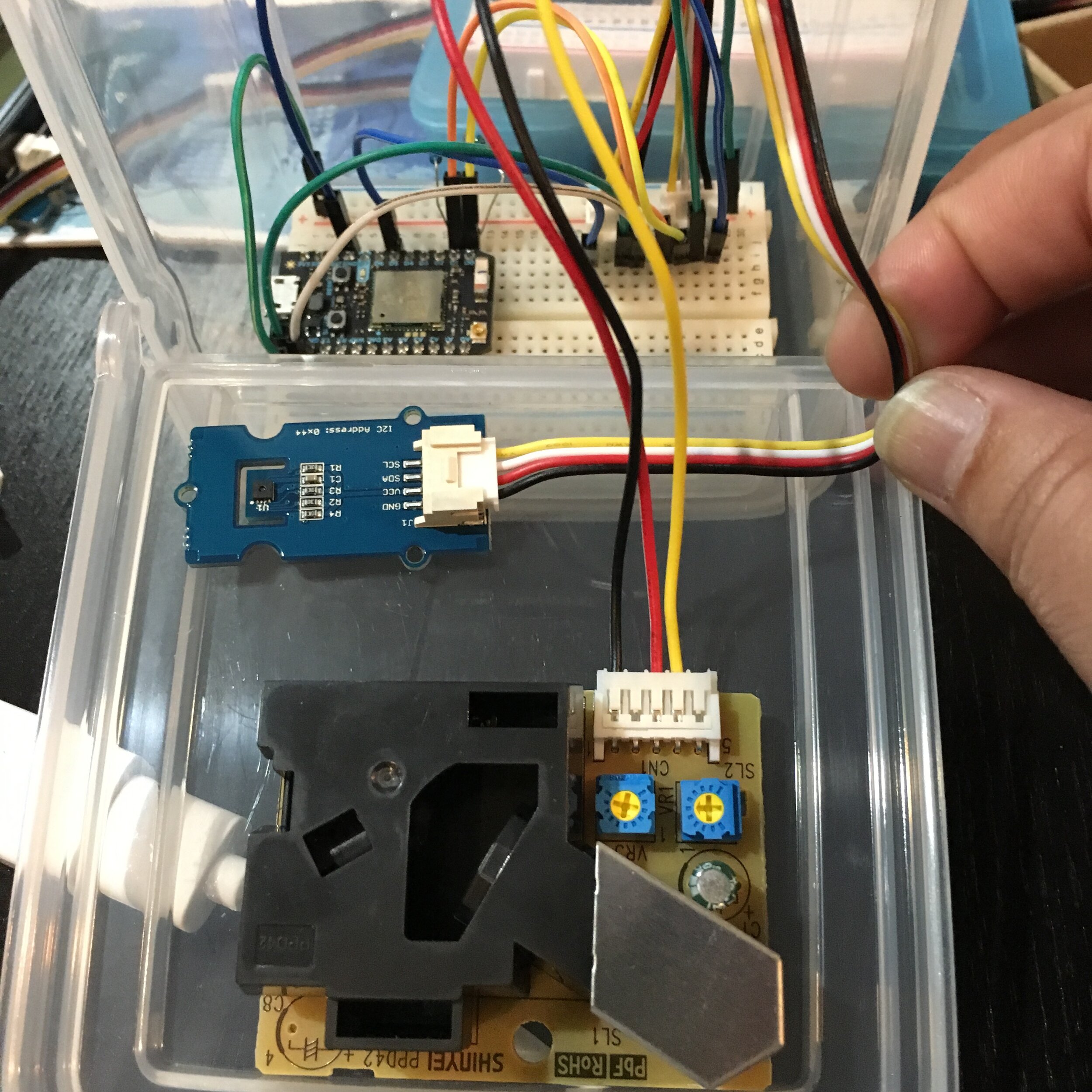
I run a URM undergraduate group at UC Merced. I focus on providing skills that students will need in real-world lab settings and train students to gain the crucial skill of disappointment management to ensure their success in the sciences. After students level up into independent projects - I help them tailor projects that fit their individual interests. For example, I helped a public health student interested in air quality build a remote particulate air sensor and program data collection to occur over wifi, pre-dental student and I developed and designed experiments for a project regarding tooth brush cleanliness and impact on oral microbiome, and worked with a pre-med student interested in childhood disease to identify the presence of antibiotic resistant bacteria in the campus pre-school classrooms.
I also lead a group that works on hunting S. aureus bacteriophages in order to create a bank for potential phage therapy uses. My program combines environmental and clinical microbiology techniques as well as bioinformatics training. Computational analysis skills are essential for today’s research and employment market - I make obtaining these skills a priority.
As a non-traditional student myself, it is important to me that I widen the ladder I scrambled up, add a ramp, and make sure it’s well-lit. I work to ensure that everyone has access to research opportunities.